Project 1: Roles for the DBHS (Drosophila Behaviour Human Splicing) family of proteins
Ermanno Moriggi, Giorgia Benegiamo
One of the projects of our lab is aimed at better understanding the physiological (and possibly pathological) roles played by a family of proteins known as DBHS (Drosophila Behaviour Human Splicing) proteins. In mammals three members belong to this family: P54NRB/NONO, PSPC1, and PSF/SFPQ, which share common structural and functional similarities (1). DBHS proteins posses two RNA recognition motifs (RRMs) and are typically labelled as “multifunctional nuclear proteins”, due to their involvement in many aspect of RNA processing, from transcriptional activation/repression (2), splicing (3), and RNA transport in neurons(4). Moreover, DBHS proteins contribute to the control of gene expression by their role (together with some non-coding RNAs) in the formation of paraspeckles, the nuclear bodies which retain within the nucleus A-I hyper-edited RNA (5) .
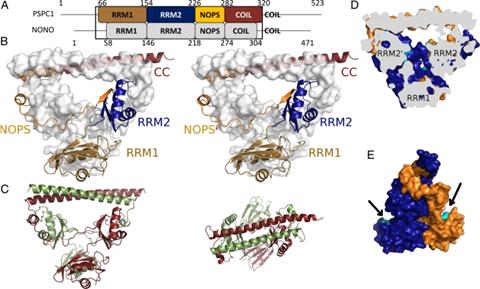
Figure 1: Structure of a PSPC1/NONO heterodimer. (A) Domain structure indicating RNA recognition motifs 1 and 2, NOPS domain, and coiled-coil. (B) Stereoview of PSPC1 (cartoon) and NONO (surface). (C) Side (Left) and top (Right) views of PSPC1 (red) and NONO (green). (D) Cutaway view revealing voids(putative RNA-binding residues, cyan). (E) Side view highlighting putative RNA-binding residues of RRM1 (cyan). Passon DM et al, PNAS, 109 :2846-50, 2012
Our laboratory first stumbled across DBHS proteins when we found that the NONO protein was important for the maintenance of mammalian circadian rhythms via association with PERIOD-protein and modulation of its transcriptional repression (6). Very recently we extended this observation to the other DBHS members, showing that they all play an important role in the circadian clock and in orchestrating circadian rhythms. In fact, by interacting with PERIOD proteins, they can bind to the promoters of many clock genes (such as RevErbPer1-2, Dbp) and modulate their repression (7).
The broad variety of functions suggests a possible role of DBHS in other aspects of circadian and non-circadian physiology. Determining the RNA and DNA interaction of DBHS proteins at the whole-genome level is essential to fully elucidate their not yet known roles in biological processes (and possibly disease states). Our lab is currying out a series of experiments to identify all the RNA and DNA targets of DBHS proteins and the consequences on the transcriptome of the presence/absence of them. These experiments combine Chromatin Immunoprecipitation (ChIP) with Next-Generation Sequencing (NGS), a new technology of sequencing.
Mouse embryonic fibroblasts from wild type and gene-trapping knock-out mice are used. ChIP in cross-linked cells allows to isolate specific RNA- or DNA-protein complexes by using antibodies against the protein of interest, providing a whole-genome picture of the protein-target interactions.
In NGS the DNA (or cDNA from RNA) obtained from ChIP is accurately sequenced in millions of parallel reactions by Illumina technology. The fragmented DNA segments are attached to a planar, optically transparent surface, then they are extended and amplified to create a high-density collection of millions short inserts (insert = a sample fragment that has been incorporated between adapters). The templates are sequenced using four-colour DNA sequencing by synthesis technology that employs reversible terminators with removable fluorescent dyes. Finally the fluorescence is detected by laser excitation and total internal reflection optics. Schematic illustrating principles of next-generation sequencing (from the BC Cancer Research Centre, Department of Breast and Molecular Oncology)
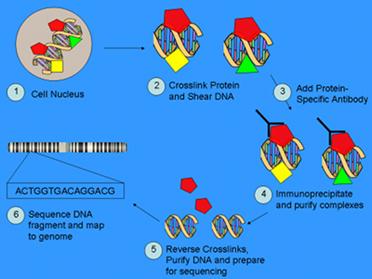
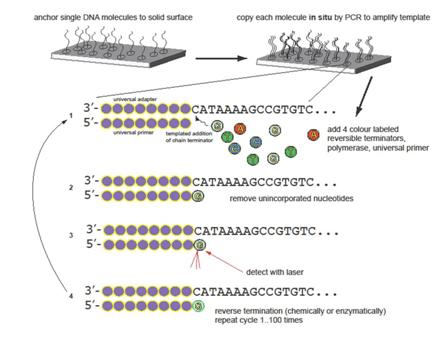
Figure 3: Schematic illustrating principles of next-generation sequencing (from the BC Cancer Research Centre, Department of Breast and Molecular Oncology)
In a first level of analysis, the identified string of bases (reads) are then mapped using a reference genome, followed by quantitation and normalization of reads to identify highly occupied binding regions.
Finally the integration of the binding regions with other information will allow to select the most relevant hits and to confirm them. The expected result will be the identification of new targets of DBHS proteins, the elucidation of their physiological roles and possible deduction of general principles of their mechanisms of action.
REFERENCES
1. Passon DM et al. 2012. Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. PNAS 109 :4846-4850
2. Ishitani K, et al. 2003. p54nrb acts as a transcriptional coactivator for activation function 1 of the human androgen receptor. Biochem. Biophys. Res. Commun. 306:660–665.
3. Kameoka S, Duque P, Konarska MM. 2004. p54(nrb) associates with the 5’ splice site within large transcription/splicing complexes. EMBO J 23: 1782–1791.
4. Kanai Y et al. 2004. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43:513–525
5. Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb Perspect Biol 2010;2:a000687
6. Brown SA, et al. 2005. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science 308:693– 696.
7. Kowalska E et al. 2012. Distinct roles of DBHS family members in the circadian transcriptional feedback loop. Mol Cell Biol 32:4585-4594.